Events
Email Subscription
Subscribe to Filter
Importing into Google Calendar
In the left-hand column of the Google Calendar main view, click the arrow to the right of "Other calendars" and click "Add by URL". In the form that appears, paste in the URL from the box above, and click the button to confirm.
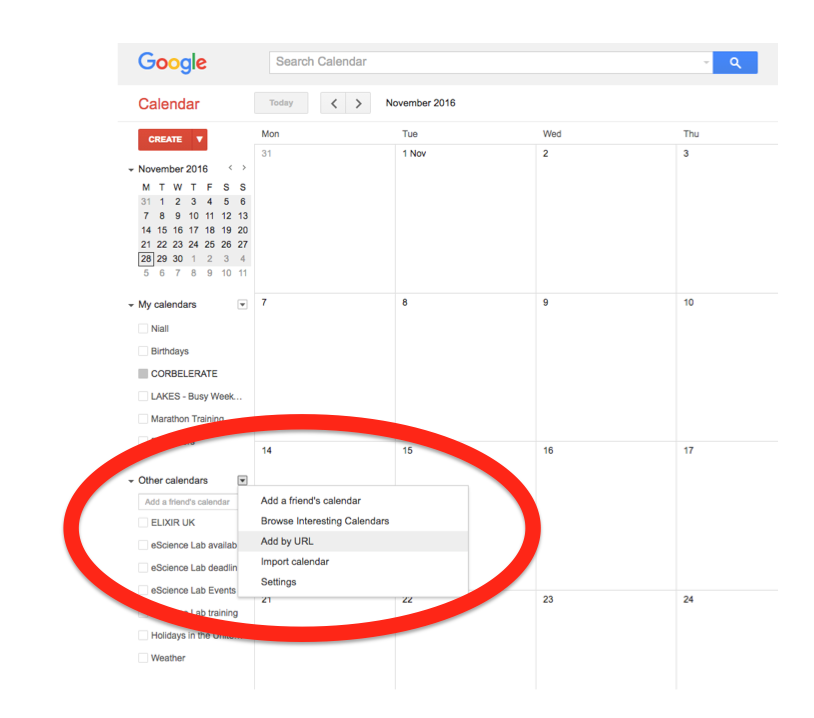
Please note, it may take a while for newly created events in TeSS to synchronise with your Google Calendar.
Content provider: European Bioinformatics Institute (EBI)
and Keywords: eQTL Catalogue
and Include expired: true
-
OnlineWorkshops and coursesUser experience design for more user-friendly applications
27 June 2017
Online
Visualisation Data quality management Fermentation Microbial ecosystems webinar UniProt: The Universal Protein Resource Proteins (proteins) BLAST Open Targets Platform Cross domain (cross-domain) Chemical biology (chemical-biology) Drug discovery Drug target identification UniRule ARBA Automated annotation MetaboLights: Metabolomics repository and reference database Chemical Entities of Biological Interest ChEBI Metabolites Molecular building blocks of life Human Cell Atlas Data Coordination Platform Single-cell transcriptomics HCA data portal Programmatic access API Python Complex Portal macromolecular assembly InterPro Boolean modelling Europe PubMed Central Literature (literature) Open access Protein Data Bank in Europe - Knowledge Base 3D structure AlphaFold Database DeepMind Artificial intelligence AI Structure prediction cancer Boolean Ensembl Genomes DNA & RNA (dna-rna) European Nucleotide Archive Data archive Raw sequencing data RNAcentral Non-coding RNA ncRNA GPU Data protection Job dispatcher Bioimage analysis resource Accessibility Missense variation Biostatistics Rfam non-coding RNA Infernal software Sequence annotation Root microbiome Abiotic stress land management Plant genotype Plant webinar series HPC database development cross-linked databases Plant database data infrastructure Plant breeding Data standards data managemnet data sharing Hyb-Seq method Flowering plants Crop improvement Pangenomics Pangenomes Virtual humans Drug-target identification plant-microbe interactions Spatial transcriptomics Plant research Drug targets Machine learning Mathematical modelling plant science Data integration plant-environment interaction Phenotyping field phenotyping Deep phenotyping EOSC-Life NHGRI-EBI GWAS Catalog clinical data genome-wide association Proteomics Proteomes Peptide search plants European Variation Archive EVA Variant clusters Variant data annotation Constraint-based metabolic modelling UniProt knowledgebase protein variant impact disease-associated protein variants Bioethics FAIR principles ELSI cohort data translational research BioModels database Mathematical modeling Reproducibity Systems biology models workflows federated analysis polygenic risk scores IntAct Molecular Interaction Database PSICQUIC IMEx Complex portal Agent-based modelling Macrophages Tumorigenesis Ensembl Training (Training) On-demand teaching introduction Building blocks Data analysis COSMIC Cancer mutation Somatic mutation ChEMBL: Bioactive data for drug discovery Chemical compounds drug-like molecules Chemogenomics Biocurator Programming Data management Green Algorithms Open data Environmental impact Carbon footprint HPC workflows Orchestrator Gene expression (gene-expression) Chemosensitivity assay Experimental protocols Drug screening MICHA European Genome-phenome Archive EGA restricted access UniProt Introduction UniProtKB Proteome Protein Data Bank in Europe genes Introductory GDPR Data security Expression Atlas Pfam protein sequence search Domain architecture Protein taxonomy AI structure prediction COVID-19 Coronavirus COVID-19 Data Portal Virology Computational simulations Signalling prior knowledge Cancer therapy response PRIDE: The Proteomics Identifications Database protein structure prediction AI system Competency framework Publication Journal club Preprints Aggregated view Structural similarity PPI networks Cell signaling Malaria Biological networks Graph theory Building biological networks Biocuration Information Manager Industry Rare-variant Variant association SKAT Sensitive data Human cohort data FAIR Gene Ontology International Mouse Phenotyping Consortium Portal Biocurators biocuration programming skills training career development Cell-level simulations Personalised medicine PerMedCoE Single cell Phenotype Genome-Wide Association Studies Genome Signaling networks BioImage Archive REST Funding UniChem: Chemical Structure Cross-referencing Bioactivity data Drug-like compounds BioStudies Database EBI Search Finding data World Sight Day Eye development Eye disease Open software Metadata Web Services Dbfetch Database fetch programmatic access Open Targets Protein structure prediction Colab notebooks RESTful API RESTful Web Services PDBe Ligand binding sites Protein families Protein functional analysis Protein sequences Data curation Biomolecular sequences Cytoscape PsyArXiv Psychology Bioactive data Drug-like molecules Biological studies Literature Neurodegenerative disorders Data sharing Molecular visualisation Molecular structures Electron density maps EBI BioSamples Database BioSamples Data Use Ontology DUO ELIXIR Ensembl Variant Effect Predictor Variant Effect Predictor VEP Variation data Annotation Macromolecular complexes UniProt Align UniProt BLAST Sequence analysis tools Multiple sequence alignments SciLite annotation Text mining MGnify Microbiome Environment Variant data Publications Citations UniProt Disease Portal ENA Sequence Search Reactome pathways database ReactomeGSA Pathway analysis Single cell RNA-seq Multiomics Data visualisation GitHub Graph database protein Gene function Mouse models IMPC Mammalian Phenotype Ontology Mouse knockouts Xenograft models Cancer EurOPDX InterProScan PDBe-KB Interpro API Europe PMC PubMed Protein function Preprints SARS-CoV-2 Genome browser Vertebrate genomes Comparative genomics Drug-like properties eQTL Catalogue Galaxy Single Cell Expression Atlas Drop-seq data SCEA PDX UniParc UniRef Genes GENCODE Gene annotation Clinical genomics Genome3D Genome annotations Protein domain prediction Gene expression Differential expression Gene regulation Alzheimer's Disease Variant and disease Enzyme annotation Rhea ENA Open-access data archive WormBase Worm genomics Parasite data Wheat genome Wheat genes QuickGO Ontologies (ontologies) Gene ontology Gene ontology annotation Controlled vocabulary Designing scientific figures Mouse genome Mouse strains Homology and variation LD calculator Linkage Disequilibrium between variants UX design UX tools user journey -
OnlineWorkshops and coursesData visualisation 101: A practical introduction to designing scientific figures
26 September 2018
Online
Visualisation Scientific communication Scientific presentation Data presentation Research presentation Protein Data Bank in Europe Ensembl UniProt: The Universal Protein Resource AlphaFold Database Proteins (proteins) DNA & RNA (dna-rna) Variant prediction Fermentation Microbial ecosystems webinar Antimicrobial resistance BLAST Open Targets Platform Cross domain (cross-domain) Chemical biology (chemical-biology) Drug discovery Drug target identification UniRule ARBA Automated annotation MetaboLights: Metabolomics repository and reference database Chemical Entities of Biological Interest ChEBI Metabolites Molecular building blocks of life Human Cell Atlas Data Coordination Platform Single-cell transcriptomics HCA data portal Programmatic access API Python Complex Portal macromolecular assembly InterPro Boolean modelling Europe PubMed Central Literature (literature) Open access Protein Data Bank in Europe - Knowledge Base 3D structure DeepMind Artificial intelligence AI Structure prediction cancer Boolean Ensembl Genomes European Nucleotide Archive Data archive Raw sequencing data RNAcentral Non-coding RNA ncRNA GPU Data protection Job dispatcher Bioimage analysis resource Accessibility Missense variation Biostatistics Rfam non-coding RNA Infernal software Sequence annotation Root microbiome Abiotic stress land management Plant genotype Plant webinar series HPC database development cross-linked databases Plant database data infrastructure Plant breeding Data standards data managemnet data sharing Hyb-Seq method Flowering plants Crop improvement Pangenomics Pangenomes Virtual humans Drug-target identification plant-microbe interactions Spatial transcriptomics Plant research Drug targets Machine learning Mathematical modelling plant science Data integration plant-environment interaction Phenotyping field phenotyping Deep phenotyping EOSC-Life NHGRI-EBI GWAS Catalog clinical data genome-wide association plants European Variation Archive EVA Variant clusters Variant data annotation Constraint-based metabolic modelling UniProt knowledgebase protein variant impact disease-associated protein variants Bioethics FAIR principles ELSI cohort data translational research BioModels database Mathematical modeling Reproducibity Systems biology models workflows federated analysis polygenic risk scores IntAct Molecular Interaction Database PSICQUIC IMEx Complex portal Agent-based modelling Macrophages Tumorigenesis Training (Training) On-demand teaching introduction Building blocks Data analysis COSMIC Cancer mutation Somatic mutation ChEMBL: Bioactive data for drug discovery Chemical compounds drug-like molecules Chemogenomics Biocurator Programming Data management Green Algorithms Open data Environmental impact Carbon footprint HPC workflows Orchestrator Gene expression (gene-expression) Chemosensitivity assay Experimental protocols Drug screening MICHA European Genome-phenome Archive EGA restricted access UniProt Introduction UniProtKB Proteome genes Introductory GDPR Data security Expression Atlas Pfam protein sequence search Domain architecture Protein taxonomy AI structure prediction COVID-19 Coronavirus COVID-19 Data Portal Virology Computational simulations Signalling prior knowledge Cancer therapy response PRIDE: The Proteomics Identifications Database protein structure prediction AI system Competency framework Publication Journal club Preprints Aggregated view Structural similarity PPI networks Cell signaling Malaria Biological networks Graph theory Building biological networks Biocuration Information Manager Industry Rare-variant Variant association SKAT Sensitive data Human cohort data FAIR Gene Ontology International Mouse Phenotyping Consortium Portal Biocurators biocuration programming skills training career development Cell-level simulations Personalised medicine PerMedCoE Single cell Phenotype Genome-Wide Association Studies Genome Signaling networks BioImage Archive REST Funding UniChem: Chemical Structure Cross-referencing Bioactivity data Drug-like compounds BioStudies Database EBI Search Finding data World Sight Day Eye development Eye disease Open software Metadata Web Services Dbfetch Database fetch programmatic access Open Targets Protein structure prediction Colab notebooks RESTful API RESTful Web Services PDBe Ligand binding sites Protein families Protein functional analysis Protein sequences Data curation Biomolecular sequences Cytoscape PsyArXiv Psychology Bioactive data Drug-like molecules Biological studies Literature Neurodegenerative disorders Data sharing Molecular visualisation Molecular structures Electron density maps EBI BioSamples Database BioSamples Data Use Ontology DUO ELIXIR Ensembl Variant Effect Predictor Variant Effect Predictor VEP Variation data Annotation SciLite annotation Text mining MGnify Microbiome Environment Proteomes Variant data Publications Citations UniProt Disease Portal ENA Sequence Search Reactome pathways database ReactomeGSA Pathway analysis Single cell RNA-seq Multiomics Data visualisation GitHub Graph database protein Gene function Mouse models IMPC Mammalian Phenotype Ontology Mouse knockouts Xenograft models Cancer EurOPDX InterProScan PDBe-KB Interpro API Europe PMC PubMed Protein function Preprints SARS-CoV-2 Genome browser Vertebrate genomes Comparative genomics Drug-like properties eQTL Catalogue Single Cell Expression Atlas SCEA Life cell by cell Introduction to single cell expression atlas PDX UniParc UniRef Genes GENCODE Gene annotation Clinical genomics Genome3D Genome annotations Protein domain prediction Gene expression Differential expression Gene regulation Alzheimer's Disease Variant and disease Enzyme annotation Rhea ENA Open-access data archive WormBase Worm genomics Parasite data Wheat genome Wheat genes QuickGO Ontologies (ontologies) Gene ontology Gene ontology annotation Controlled vocabulary Designing scientific figures -
OnlineWorkshops and coursesHMMER: Fast and sensitive sequence similarity searches
14 November 2018
Online
Proteins Sequence analysis Sequence search results Open Targets Platform Cross domain (cross-domain) Chemical biology (chemical-biology) Drug discovery Drug target identification UniProt: The Universal Protein Resource Proteins (proteins) UniRule ARBA Automated annotation MetaboLights: Metabolomics repository and reference database Chemical Entities of Biological Interest ChEBI Metabolites Molecular building blocks of life Human Cell Atlas Data Coordination Platform Single-cell transcriptomics HCA data portal Programmatic access API Python Complex Portal macromolecular assembly InterPro Boolean modelling Europe PubMed Central Literature (literature) Open access Protein Data Bank in Europe - Knowledge Base 3D structure AlphaFold Database DeepMind Artificial intelligence AI Structure prediction cancer Boolean Ensembl Genomes DNA & RNA (dna-rna) European Nucleotide Archive Data archive Raw sequencing data RNAcentral Non-coding RNA ncRNA GPU Data protection Job dispatcher Bioimage analysis resource Accessibility Missense variation Biostatistics Rfam non-coding RNA Infernal software Sequence annotation Root microbiome Abiotic stress land management Plant genotype Plant webinar series HPC database development cross-linked databases Plant database data infrastructure Plant breeding Data standards data managemnet data sharing Hyb-Seq method Flowering plants Crop improvement Pangenomics Pangenomes Virtual humans Drug-target identification plant-microbe interactions Spatial transcriptomics Plant research Drug targets Machine learning Mathematical modelling plant science Data integration plant-environment interaction Phenotyping field phenotyping Deep phenotyping EOSC-Life NHGRI-EBI GWAS Catalog clinical data genome-wide association Proteomics Proteomes Peptide search plants European Variation Archive EVA Variant clusters Variant data annotation Constraint-based metabolic modelling UniProt knowledgebase protein variant impact disease-associated protein variants Bioethics FAIR principles ELSI cohort data translational research BioModels database Mathematical modeling Reproducibity Systems biology models workflows federated analysis polygenic risk scores IntAct Molecular Interaction Database PSICQUIC IMEx Complex portal Agent-based modelling Macrophages Tumorigenesis Ensembl Training (Training) On-demand teaching introduction Building blocks Data analysis COSMIC Cancer mutation Somatic mutation UniFIRE Protein annotation ChEMBL: Bioactive data for drug discovery Chemical compounds drug-like molecules Chemogenomics Biocurator Programming Data management Green Algorithms Open data Environmental impact Carbon footprint HPC workflows Orchestrator Gene expression (gene-expression) Chemosensitivity assay Experimental protocols Drug screening MICHA European Genome-phenome Archive EGA restricted access UniProt Introduction UniProtKB Proteome Protein Data Bank in Europe genes Introductory GDPR Data security Expression Atlas Pfam protein sequence search Domain architecture Protein taxonomy Beginner AI structure prediction COVID-19 Coronavirus COVID-19 Data Portal Virology Computational simulations Signalling prior knowledge Cancer therapy response PRIDE: The Proteomics Identifications Database protein structure prediction AI system Competency framework Publication Journal club Preprints Aggregated view Structural similarity PPI networks Cell signaling Malaria Biological networks Graph theory Building biological networks Biocuration Information Manager Industry Rare-variant Variant association SKAT Sensitive data Human cohort data FAIR Gene Ontology International Mouse Phenotyping Consortium Portal Biocurators biocuration programming skills training career development Cell-level simulations Personalised medicine PerMedCoE Single cell Phenotype Genome-Wide Association Studies Genome Signaling networks BioImage Archive REST Funding UniChem: Chemical Structure Cross-referencing Bioactivity data Drug-like compounds BioStudies Database EBI Search Finding data World Sight Day Eye development Eye disease Open software Metadata Web Services Dbfetch Database fetch programmatic access Open Targets Protein structure prediction Colab notebooks RESTful API RESTful Web Services PDBe Ligand binding sites Protein families Protein functional analysis Protein sequences Data curation Biomolecular sequences Reactome pathways database ReactomeGSA scRNA-seq Multiomics Cytoscape PsyArXiv Psychology Bioactive data Drug-like molecules Biological studies Literature Neurodegenerative disorders RNA families Data sharing Molecular visualisation Molecular structures Electron density maps EBI BioSamples Database BioSamples Data Use Ontology DUO ELIXIR Ensembl Variant Effect Predictor Variant Effect Predictor VEP Variation data Annotation Literature search Publications Macromolecular complexes UniProt Align UniProt BLAST Sequence analysis tools Multiple sequence alignments SciLite annotation Text mining MGnify Microbiome Environment Variant data Citations UniProt Disease Portal ENA Sequence Search Pathway analysis Single cell RNA-seq Data visualisation GitHub Ensembl Rapid Release Graph database protein Gene function Mouse models IMPC Mammalian Phenotype Ontology Mouse knockouts Xenograft models Cancer EurOPDX InterProScan Enzyme Portal Metabolism PDBe-KB Interpro API Europe PMC PubMed Protein function Preprints SARS-CoV-2 Genome browser Vertebrate genomes Comparative genomics Drug-like properties eQTL Catalogue Galaxy Single Cell Expression Atlas Drop-seq data SCEA PDX UniParc UniRef Genes GENCODE Gene annotation Clinical genomics Genome3D Genome annotations Protein domain prediction Functional annotation UniFire Gene expression Differential expression Gene regulation Enzyme-related information Alzheimer's Disease Variant and disease Enzyme annotation Rhea ENA Open-access data archive WormBase Worm genomics Parasite data Wheat genome Wheat genes HMMER - protein homology search Hidden Markov models Sequence similarity search Protein homology -
OnlineWorkshops and coursesEnzyme Portal
29 May 2019
Online
Enzymes Open Targets Platform Cross domain (cross-domain) Chemical biology (chemical-biology) Drug discovery Drug target identification UniProt: The Universal Protein Resource Proteins (proteins) UniRule ARBA Automated annotation MetaboLights: Metabolomics repository and reference database Chemical Entities of Biological Interest ChEBI Metabolites Molecular building blocks of life Human Cell Atlas Data Coordination Platform Single-cell transcriptomics HCA data portal Programmatic access API Python Complex Portal macromolecular assembly InterPro Boolean modelling Europe PubMed Central Literature (literature) Open access Protein Data Bank in Europe - Knowledge Base 3D structure AlphaFold Database DeepMind Artificial intelligence AI Structure prediction cancer Boolean Ensembl Genomes DNA & RNA (dna-rna) European Nucleotide Archive Data archive Raw sequencing data RNAcentral Non-coding RNA ncRNA GPU Data protection Job dispatcher Bioimage analysis resource Accessibility Missense variation Biostatistics Rfam non-coding RNA Infernal software Sequence annotation Root microbiome Abiotic stress land management Plant genotype Plant webinar series HPC database development cross-linked databases Plant database data infrastructure Plant breeding Data standards data managemnet data sharing Hyb-Seq method Flowering plants Crop improvement Pangenomics Pangenomes Virtual humans Drug-target identification plant-microbe interactions Spatial transcriptomics Plant research Drug targets Machine learning Mathematical modelling plant science Data integration plant-environment interaction Phenotyping field phenotyping Deep phenotyping EOSC-Life NHGRI-EBI GWAS Catalog clinical data genome-wide association Proteomics Proteomes Peptide search plants European Variation Archive EVA Variant clusters Variant data annotation Constraint-based metabolic modelling UniProt knowledgebase protein variant impact disease-associated protein variants Bioethics FAIR principles ELSI cohort data translational research BioModels database Mathematical modeling Reproducibity Systems biology models workflows federated analysis polygenic risk scores IntAct Molecular Interaction Database PSICQUIC IMEx Complex portal Agent-based modelling Macrophages Tumorigenesis Ensembl Training (Training) On-demand teaching introduction Building blocks Data analysis COSMIC Cancer mutation Somatic mutation UniFIRE Protein annotation ChEMBL: Bioactive data for drug discovery Chemical compounds drug-like molecules Chemogenomics Biocurator Programming Data management Green Algorithms Open data Environmental impact Carbon footprint HPC workflows Orchestrator Gene expression (gene-expression) Chemosensitivity assay Experimental protocols Drug screening MICHA European Genome-phenome Archive EGA restricted access UniProt Introduction UniProtKB Proteome Protein Data Bank in Europe genes Introductory GDPR Data security Expression Atlas Pfam protein sequence search Domain architecture Protein taxonomy Beginner AI structure prediction COVID-19 Coronavirus COVID-19 Data Portal Virology Computational simulations Signalling prior knowledge Cancer therapy response PRIDE: The Proteomics Identifications Database protein structure prediction AI system Competency framework Publication Journal club Preprints Aggregated view Structural similarity PPI networks Cell signaling Malaria Biological networks Graph theory Building biological networks Biocuration Information Manager Industry Rare-variant Variant association SKAT Sensitive data Human cohort data FAIR Gene Ontology International Mouse Phenotyping Consortium Portal Biocurators biocuration programming skills training career development Cell-level simulations Personalised medicine PerMedCoE Single cell Phenotype Genome-Wide Association Studies Genome Signaling networks BioImage Archive REST Funding UniChem: Chemical Structure Cross-referencing Bioactivity data Drug-like compounds BioStudies Database EBI Search Finding data World Sight Day Eye development Eye disease Open software Metadata Web Services Dbfetch Database fetch programmatic access Open Targets Protein structure prediction Colab notebooks RESTful API RESTful Web Services PDBe Ligand binding sites Protein families Protein functional analysis Protein sequences Data curation Biomolecular sequences Reactome pathways database ReactomeGSA scRNA-seq Multiomics Cytoscape PsyArXiv Psychology Bioactive data Drug-like molecules Biological studies Literature Neurodegenerative disorders RNA families Data sharing Molecular visualisation Molecular structures Electron density maps EBI BioSamples Database BioSamples Data Use Ontology DUO ELIXIR Ensembl Variant Effect Predictor Variant Effect Predictor VEP Variation data Annotation Literature search Publications Macromolecular complexes UniProt Align UniProt BLAST Sequence analysis tools Multiple sequence alignments SciLite annotation Text mining MGnify Microbiome Environment Variant data Citations UniProt Disease Portal ENA Sequence Search Pathway analysis Single cell RNA-seq Data visualisation GitHub Ensembl Rapid Release Graph database protein Gene function Mouse models IMPC Mammalian Phenotype Ontology Mouse knockouts Xenograft models Cancer EurOPDX InterProScan Enzyme Portal Metabolism PDBe-KB Interpro API Europe PMC PubMed Protein function Preprints SARS-CoV-2 Genome browser Vertebrate genomes Comparative genomics Drug-like properties eQTL Catalogue Galaxy Single Cell Expression Atlas Drop-seq data SCEA PDX UniParc UniRef Genes GENCODE Gene annotation Clinical genomics Genome3D Genome annotations Protein domain prediction Functional annotation UniFire Gene expression Differential expression Gene regulation Enzyme-related information -
OnlineWorkshops and coursesAnnotate your proteins with the UniProt Functional Annotation system UniFire
10 July 2019
Online
Proteins Protein function prediction Open Targets Platform Cross domain (cross-domain) Chemical biology (chemical-biology) Drug discovery Drug target identification UniProt: The Universal Protein Resource Proteins (proteins) UniRule ARBA Automated annotation MetaboLights: Metabolomics repository and reference database Chemical Entities of Biological Interest ChEBI Metabolites Molecular building blocks of life Human Cell Atlas Data Coordination Platform Single-cell transcriptomics HCA data portal Programmatic access API Python Complex Portal macromolecular assembly InterPro Boolean modelling Europe PubMed Central Literature (literature) Open access Protein Data Bank in Europe - Knowledge Base 3D structure AlphaFold Database DeepMind Artificial intelligence AI Structure prediction cancer Boolean Ensembl Genomes DNA & RNA (dna-rna) European Nucleotide Archive Data archive Raw sequencing data RNAcentral Non-coding RNA ncRNA GPU Data protection Job dispatcher Bioimage analysis resource Accessibility Missense variation Biostatistics Rfam non-coding RNA Infernal software Sequence annotation Root microbiome Abiotic stress land management Plant genotype Plant webinar series HPC database development cross-linked databases Plant database data infrastructure Plant breeding Data standards data managemnet data sharing Hyb-Seq method Flowering plants Crop improvement Pangenomics Pangenomes Virtual humans Drug-target identification plant-microbe interactions Spatial transcriptomics Plant research Drug targets Machine learning Mathematical modelling plant science Data integration plant-environment interaction Phenotyping field phenotyping Deep phenotyping EOSC-Life NHGRI-EBI GWAS Catalog clinical data genome-wide association Proteomics Proteomes Peptide search plants European Variation Archive EVA Variant clusters Variant data annotation Constraint-based metabolic modelling UniProt knowledgebase protein variant impact disease-associated protein variants Bioethics FAIR principles ELSI cohort data translational research BioModels database Mathematical modeling Reproducibity Systems biology models workflows federated analysis polygenic risk scores IntAct Molecular Interaction Database PSICQUIC IMEx Complex portal Agent-based modelling Macrophages Tumorigenesis Ensembl Training (Training) On-demand teaching introduction Building blocks Data analysis COSMIC Cancer mutation Somatic mutation UniFIRE Protein annotation ChEMBL: Bioactive data for drug discovery Chemical compounds drug-like molecules Chemogenomics Biocurator Programming Data management Green Algorithms Open data Environmental impact Carbon footprint HPC workflows Orchestrator Gene expression (gene-expression) Chemosensitivity assay Experimental protocols Drug screening MICHA European Genome-phenome Archive EGA restricted access UniProt Introduction UniProtKB Proteome Protein Data Bank in Europe genes Introductory GDPR Data security Expression Atlas Pfam protein sequence search Domain architecture Protein taxonomy Beginner AI structure prediction COVID-19 Coronavirus COVID-19 Data Portal Virology Computational simulations Signalling prior knowledge Cancer therapy response PRIDE: The Proteomics Identifications Database protein structure prediction AI system Competency framework Publication Journal club Preprints Aggregated view Structural similarity PPI networks Cell signaling Malaria Biological networks Graph theory Building biological networks Biocuration Information Manager Industry Rare-variant Variant association SKAT Sensitive data Human cohort data FAIR Gene Ontology International Mouse Phenotyping Consortium Portal Biocurators biocuration programming skills training career development Cell-level simulations Personalised medicine PerMedCoE Single cell Phenotype Genome-Wide Association Studies Genome Signaling networks BioImage Archive REST Funding UniChem: Chemical Structure Cross-referencing Bioactivity data Drug-like compounds BioStudies Database EBI Search Finding data World Sight Day Eye development Eye disease Open software Metadata Web Services Dbfetch Database fetch programmatic access Open Targets Protein structure prediction Colab notebooks RESTful API RESTful Web Services PDBe Ligand binding sites Protein families Protein functional analysis Protein sequences Data curation Biomolecular sequences Reactome pathways database ReactomeGSA scRNA-seq Multiomics Cytoscape PsyArXiv Psychology Bioactive data Drug-like molecules Biological studies Literature Neurodegenerative disorders RNA families Data sharing Molecular visualisation Molecular structures Electron density maps EBI BioSamples Database BioSamples Data Use Ontology DUO ELIXIR Ensembl Variant Effect Predictor Variant Effect Predictor VEP Variation data Annotation Literature search Publications Macromolecular complexes UniProt Align UniProt BLAST Sequence analysis tools Multiple sequence alignments SciLite annotation Text mining MGnify Microbiome Environment Variant data Citations UniProt Disease Portal ENA Sequence Search Pathway analysis Single cell RNA-seq Data visualisation GitHub Ensembl Rapid Release Graph database protein Gene function Mouse models IMPC Mammalian Phenotype Ontology Mouse knockouts Xenograft models Cancer EurOPDX InterProScan Enzyme Portal Metabolism PDBe-KB Interpro API Europe PMC PubMed Protein function Preprints SARS-CoV-2 Genome browser Vertebrate genomes Comparative genomics Drug-like properties eQTL Catalogue Galaxy Single Cell Expression Atlas Drop-seq data SCEA PDX UniParc UniRef Genes GENCODE Gene annotation Clinical genomics Genome3D Genome annotations Protein domain prediction Functional annotation UniFire -
OnlineWorkshops and coursesSingle cell RNA-seq analysis using a Galaxy interface
11 December 2019
Online
RNA-Seq DNA & RNA (dna-rna) Proteins (proteins) UniProt: The Universal Protein Resource Fermentation Microbial ecosystems webinar Antimicrobial resistance Ensembl BLAST Open Targets Platform Cross domain (cross-domain) Chemical biology (chemical-biology) Drug discovery Drug target identification UniRule ARBA Automated annotation MetaboLights: Metabolomics repository and reference database Chemical Entities of Biological Interest ChEBI Metabolites Molecular building blocks of life Human Cell Atlas Data Coordination Platform Single-cell transcriptomics HCA data portal Programmatic access API Python Complex Portal macromolecular assembly InterPro Boolean modelling Europe PubMed Central Literature (literature) Open access Protein Data Bank in Europe - Knowledge Base 3D structure AlphaFold Database DeepMind Artificial intelligence AI Structure prediction cancer Boolean Ensembl Genomes European Nucleotide Archive Data archive Raw sequencing data RNAcentral Non-coding RNA ncRNA GPU Data protection Job dispatcher Bioimage analysis resource Accessibility Missense variation Biostatistics Rfam non-coding RNA Infernal software Sequence annotation Root microbiome Abiotic stress land management Plant genotype Plant webinar series HPC database development cross-linked databases Plant database data infrastructure Plant breeding Data standards data managemnet data sharing Hyb-Seq method Flowering plants Crop improvement Pangenomics Pangenomes Virtual humans Drug-target identification plant-microbe interactions Spatial transcriptomics Plant research Drug targets Machine learning Mathematical modelling plant science Data integration plant-environment interaction Phenotyping field phenotyping Deep phenotyping EOSC-Life NHGRI-EBI GWAS Catalog clinical data genome-wide association plants European Variation Archive EVA Variant clusters Variant data annotation Constraint-based metabolic modelling UniProt knowledgebase protein variant impact disease-associated protein variants Bioethics FAIR principles ELSI cohort data translational research BioModels database Mathematical modeling Reproducibity Systems biology models workflows federated analysis polygenic risk scores IntAct Molecular Interaction Database PSICQUIC IMEx Complex portal Agent-based modelling Macrophages Tumorigenesis Training (Training) On-demand teaching introduction Building blocks Data analysis COSMIC Cancer mutation Somatic mutation ChEMBL: Bioactive data for drug discovery Chemical compounds drug-like molecules Chemogenomics Biocurator Programming Data management Green Algorithms Open data Environmental impact Carbon footprint HPC workflows Orchestrator Gene expression (gene-expression) Chemosensitivity assay Experimental protocols Drug screening MICHA European Genome-phenome Archive EGA restricted access UniProt Introduction UniProtKB Proteome Protein Data Bank in Europe genes Introductory GDPR Data security Expression Atlas Pfam protein sequence search Domain architecture Protein taxonomy AI structure prediction COVID-19 Coronavirus COVID-19 Data Portal Virology Computational simulations Signalling prior knowledge Cancer therapy response PRIDE: The Proteomics Identifications Database protein structure prediction AI system Competency framework Publication Journal club Preprints Aggregated view Structural similarity PPI networks Cell signaling Malaria Biological networks Graph theory Building biological networks Biocuration Information Manager Industry Rare-variant Variant association SKAT Sensitive data Human cohort data FAIR Gene Ontology International Mouse Phenotyping Consortium Portal Biocurators biocuration programming skills training career development Cell-level simulations Personalised medicine PerMedCoE Single cell Phenotype Genome-Wide Association Studies Genome Signaling networks BioImage Archive REST Funding UniChem: Chemical Structure Cross-referencing Bioactivity data Drug-like compounds BioStudies Database EBI Search Finding data World Sight Day Eye development Eye disease Open software Metadata Web Services Dbfetch Database fetch programmatic access Open Targets Protein structure prediction Colab notebooks RESTful API RESTful Web Services PDBe Ligand binding sites Protein families Protein functional analysis Protein sequences Data curation Biomolecular sequences Cytoscape PsyArXiv Psychology Bioactive data Drug-like molecules Biological studies Literature Neurodegenerative disorders Data sharing Molecular visualisation Molecular structures Electron density maps EBI BioSamples Database BioSamples Data Use Ontology DUO ELIXIR Ensembl Variant Effect Predictor Variant Effect Predictor VEP Variation data Annotation SciLite annotation Text mining MGnify Microbiome Environment Proteomes Variant data Publications Citations UniProt Disease Portal ENA Sequence Search Reactome pathways database ReactomeGSA Pathway analysis Single cell RNA-seq Multiomics Data visualisation GitHub Graph database protein Gene function Mouse models IMPC Mammalian Phenotype Ontology Mouse knockouts Xenograft models Cancer EurOPDX InterProScan PDBe-KB Interpro API Europe PMC PubMed Protein function Preprints SARS-CoV-2 Genome browser Vertebrate genomes Comparative genomics Drug-like properties eQTL Catalogue Galaxy Single Cell Expression Atlas Drop-seq data -
OnlineWorkshops and courseseQTL Catalogue: a compendium of uniformly processed human expression and splicing QTLs
12 February 2020 @ 09:00 - 17:00
Online
GWAS study Genetic variation Gene expression QTL analysis Open Targets eQTL Catalogue API -
OnlineWorkshops and coursesAdvances in spatial omics
11 September - 16 October 2024
Online
Data integration Advances in spatial omics RNA FISH Spatial multi-omics BioImage Archive Single cell Spatial transcriptomics Tissue biology Cell-cell interactions Cell signalling cell-cell communication cell-cell interaction single-cells Scientific communication Scientific presentation Data presentation Research presentation Protein Data Bank in Europe Ensembl UniProt: The Universal Protein Resource AlphaFold Database Proteins (proteins) DNA & RNA (dna-rna) Variant prediction Fermentation Microbial ecosystems webinar Antimicrobial resistance BLAST Open Targets Platform Cross domain (cross-domain) Chemical biology (chemical-biology) Drug discovery Drug target identification UniRule ARBA Automated annotation MetaboLights: Metabolomics repository and reference database Chemical Entities of Biological Interest ChEBI Metabolites Molecular building blocks of life Human Cell Atlas Data Coordination Platform Single-cell transcriptomics HCA data portal Programmatic access API Python Complex Portal macromolecular assembly InterPro Boolean modelling Europe PubMed Central Literature (literature) Open access Protein Data Bank in Europe - Knowledge Base 3D structure DeepMind Artificial intelligence AI Structure prediction cancer Boolean Ensembl Genomes European Nucleotide Archive Data archive Raw sequencing data RNAcentral Non-coding RNA ncRNA GPU Data protection Job dispatcher Bioimage analysis resource Accessibility Missense variation Biostatistics Rfam non-coding RNA Infernal software Sequence annotation Root microbiome Abiotic stress land management Plant genotype Plant webinar series HPC database development cross-linked databases Plant database data infrastructure Plant breeding Data standards data managemnet data sharing Hyb-Seq method Flowering plants Crop improvement Pangenomics Pangenomes Virtual humans Drug-target identification plant-microbe interactions Plant research Drug targets Machine learning Mathematical modelling plant science plant-environment interaction Phenotyping field phenotyping Deep phenotyping EOSC-Life NHGRI-EBI GWAS Catalog clinical data genome-wide association plants European Variation Archive EVA Variant clusters Variant data annotation Constraint-based metabolic modelling UniProt knowledgebase protein variant impact disease-associated protein variants Bioethics FAIR principles ELSI cohort data translational research BioModels database Mathematical modeling Reproducibity Systems biology models workflows federated analysis polygenic risk scores IntAct Molecular Interaction Database PSICQUIC IMEx Complex portal Agent-based modelling Macrophages Tumorigenesis Training (Training) On-demand teaching introduction Building blocks Data analysis COSMIC Cancer mutation Somatic mutation ChEMBL: Bioactive data for drug discovery Chemical compounds drug-like molecules Chemogenomics Biocurator Programming Data management Green Algorithms Open data Environmental impact Carbon footprint HPC workflows Orchestrator Gene expression (gene-expression) Chemosensitivity assay Experimental protocols Drug screening MICHA European Genome-phenome Archive EGA restricted access UniProt Introduction UniProtKB Proteome genes Introductory GDPR Data security Expression Atlas Pfam protein sequence search Domain architecture Protein taxonomy AI structure prediction COVID-19 Coronavirus COVID-19 Data Portal Virology Computational simulations Signalling prior knowledge Cancer therapy response PRIDE: The Proteomics Identifications Database protein structure prediction AI system Competency framework Publication Journal club Preprints Aggregated view Structural similarity PPI networks Cell signaling Malaria Biological networks Graph theory Building biological networks Biocuration Information Manager Industry Rare-variant Variant association SKAT Sensitive data Human cohort data FAIR Gene Ontology International Mouse Phenotyping Consortium Portal Biocurators biocuration programming skills training career development Cell-level simulations Personalised medicine PerMedCoE Phenotype Genome-Wide Association Studies Genome Signaling networks REST Funding UniChem: Chemical Structure Cross-referencing Bioactivity data Drug-like compounds BioStudies Database EBI Search Finding data World Sight Day Eye development Eye disease Open software Metadata Web Services Dbfetch Database fetch programmatic access Open Targets Protein structure prediction Colab notebooks RESTful API RESTful Web Services PDBe Ligand binding sites Protein families Protein functional analysis Protein sequences Data curation Biomolecular sequences Cytoscape PsyArXiv Psychology Bioactive data Drug-like molecules Biological studies Literature Neurodegenerative disorders Data sharing Molecular visualisation Molecular structures Electron density maps EBI BioSamples Database BioSamples Data Use Ontology DUO ELIXIR Ensembl Variant Effect Predictor Variant Effect Predictor VEP Variation data Annotation SciLite annotation Text mining MGnify Microbiome Environment Proteomes Variant data Publications Citations UniProt Disease Portal ENA Sequence Search Reactome pathways database ReactomeGSA Pathway analysis Single cell RNA-seq Multiomics Data visualisation GitHub Graph database protein Gene function Mouse models IMPC Mammalian Phenotype Ontology Mouse knockouts Xenograft models Cancer EurOPDX InterProScan PDBe-KB Interpro API Europe PMC PubMed Protein function Preprints SARS-CoV-2 Genome browser Vertebrate genomes Comparative genomics Drug-like properties eQTL Catalogue Single Cell Expression Atlas SCEA Life cell by cell Introduction to single cell expression atlas PDX UniParc UniRef Genes GENCODE Gene annotation Clinical genomics Genome3D Genome annotations Protein domain prediction Gene expression Differential expression Gene regulation Alzheimer's Disease Variant and disease Enzyme annotation Rhea ENA Open-access data archive WormBase Worm genomics Parasite data Wheat genome Wheat genes QuickGO Ontologies (ontologies) Gene ontology Gene ontology annotation Controlled vocabulary Mouse genome Mouse strains Homology and variation LD calculator Linkage Disequilibrium between variants
Note, this map only displays events that have geolocation information in
TeSS.
For the complete list of events in TeSS, click the grid tab.